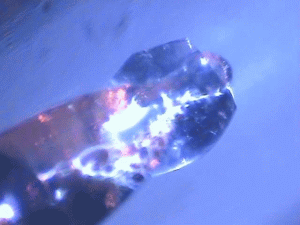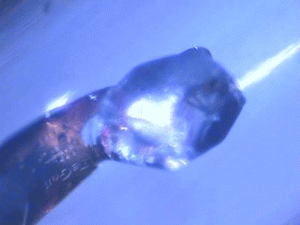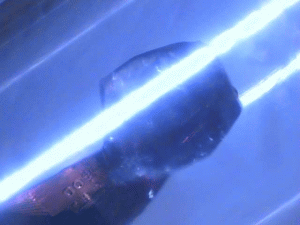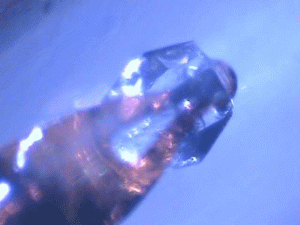
| Protein crystals are fascinating materials that provide 80% of known protein structures. These crystals form due to weak intermolecular interactions and often include 50% by volume water, including mobile water. These ordered array of biomacromolecules provide the needed orientational alignment to determine protein structure and characterize underlying protein dynamics using time resolved protein crystallography and anisotropic terahertz microspectroscopy. However, the specific intermolecular interactions perturb the measured dynamics from those in vivo. We use ATM to characterize these perturbations by comparing the measured spectra with calculated spectra for different crystal symmetry groups. These studies include protein expression, purification and crystallization. Crystal unit cell and face indexing with X-ray diffraction. Protein collective vibration spectroscopy using Anisotropic Terahertz Microspectroscopy (ATM). |
 |
|